RUNX1::RUNX1T1 Fusion in Pediatric Acute Myeloid Leukemia
A Description of Two Cases
DOI:
https://doi.org/10.21141/PJP.2023.02Keywords:
Pediatric acute myeloid leukemia, Next Generation Sequencing, RUNX1::RUNX1T1 fusion, Berlin-Frankfurt-Münster (BFM-87) protocol, AML 15 Medical Research Council protocolAbstract
RUNX1::RUNX1T1 is a core-binding factor driving fusion gene which arises from t(8;21)(q22;q22). It is one of the most common chromosomal rearrangements in both pediatric and adult Acute Myeloid Leukemia (AML) with a reported incidence of 15% in children and young adults. There are few case reports documenting RUNX1::RUNX1T1 translocation in pediatric AML. Although this is generally associated with a favorable prognosis, we report two (2) cases of de novo pediatric AML in the Philippines harboring a RUNX1::RUNX1T1 translocation, one eventually relapsed while the other attained remission but succumbed to sepsis.
Downloads
References
2. McPherson RA, Pincus M. Henry’s clinical diagnosis and management by laboratory methods. 23rd ed. Saunders Elsevier; 2016.
3. Swerdlow SH. WHO classification of tumours of haematopoietic and lymphoid tissues Revised 4th ed, vol 2. International Agency for Research on Cancer; 2017.
4. George TI, Bajel A. Diagnosis of rare subtypes of acute myeloid leukaemia and related neoplasms. Pathology. 2021;53(3):312-27. https://pubmed.ncbi.nlm.nih.gov/33676766. https://doi.org/10.1016/j.pathol.2021.02.001.
5. Tamayo SCA, Medalla IMC, Roque EMP, et al. Molecular characterization of acute myeloid leukemia among pediatric Filipino patients using karyotype analysis, fluorescence in situ hybridization and comprehensive next generation sequencing: a multi-center experience [abstract]. In: 35th International Symposium on Technical Innovations in Laboratory Hematology; 2022 Sep 8-10; Bologna, Italy: ISLH; 2022. Abstract number 93.
6. Höllein A, Nadarajah N, Meggendorfer M, et al. Molecular characterization of AML with RUNX1-RUNX1T1 at diagnosis and relapse reveals net loss of co-mutations. Hemasphere. 2019;3(1):e178. https://pubmed.ncbi.nlm.nih.gov/31723813. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6745937. https://doi.org/10.1097/HS9.0000000000000178.
7. Swart LE, Heidenreich O. The RUNX1/RUNX1T1 network: translating insights into therapeutic options. Exp Hematol. 2021;94:1-10. https://pubmed.ncbi.nlm.nih.gov/33217477. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7854360. https://doi.org/10.1016/j.exphem.2020.11.005.
8. Rasche M, Zimmermann M, Borschel L, et al. Successes and challenges in the treatment of pediatric acute myeloid leukemia: a retrospective analysis of the AML-BFM trials from 1987 to 2012. Leukemia. 2018;32(10):2167-77. https://pubmed.ncbi.nlm.nih.gov/29550834. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6170392. https://doi.org/10.1038/s41375-018-0071-7.
9. Hu Y, Chen A, Zheng X, et al. Ecological principle meets cancer treatment: treating children with acute myeloid leukemia with low-dose chemotherapy. Natl Sci Rev. 2019;6(3):469-79. https://pubmed.ncbi.nlm.nih.gov/34691895. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8291445. https://doi.org/10.1093/nsr/nwz006.
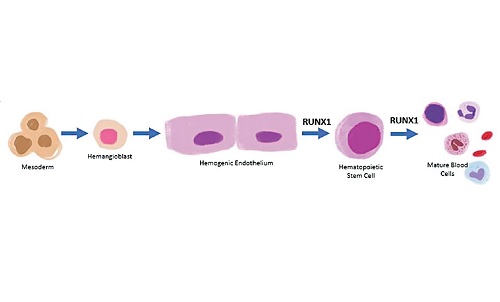
10. Gao L, Tober J, Gao P, Chen C, Tan K, Speck NA. RUNX1 and the endothelial origin of blood. Exp Hematol. 2018;68:2-9. https://pubmed.ncbi.nlm.nih.gov/30391350. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6494457. https://doi.org/10.1016/j.exphem.2018.10.009.
11. Daniel MG, Rapp K, Schaniel C, Moore KA. Induction of developmental hematopoiesis mediated by transcription factors and the hematopoietic microenvironment. Ann N Y Acad Sci. 2020;1466(1):59-72. https://pubmed.ncbi.nlm.nih.gov/31621095. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7162702. https://doi.org/10.1111/nyas.14246.
12. Krauth M-T, Eder C, Alpermann T, et al. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome. Leukemia. 2014;28(7):1449-58. https://pubmed.ncbi.nlm.nih.gov/24402164. https://doi.org/10.1038/leu.2014.4.
13. Kita K, Nakase K, Miwa H, et al. Phenotypical characteristics of acute myelocytic leukemia associated with the t(8;21)(q22;q22) chromosomal abnormality: frequent expression of immature B-cell antigen CD19 together with stem cell antigen CD34. Blood. 1992;80(2):470-7. https://pubmed.ncbi.nlm.nih.gov/1378322.
14. Haferlach T, Meggendorfer M. More than a fusion gene: the RUNX1-RUNX1T1 AML. Blood. 2019;133(10):1006-7. https://pubmed.ncbi.nlm.nih.gov/30846508. https://doi.org/10.1182/blood-2019-01-896076.
15. Aung MMK, Mills ML, Bittencourt‐Silvestre J, Keeshan K. Insights into the molecular profiles of adult and paediatric acute myeloid leukaemia. Mol Oncol. 2021;15(9):2253-72. https://pubmed.ncbi.nlm.nih.gov/33421304. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8410545. https://doi.org/10.1002/1878-0261.12899.
16. Krock B, Oberley MJ. Molecular genetics of pediatric acute myeloid leukemia. Clin Lab Med. 2021;41(3):497-515. https://pubmed.ncbi.nlm.nih.gov/34304778. https://doi.org/10.1016/j.cll.2021.03.014.
17. Reinhardt D, Antoniou E, Waack K. Pediatric acute myeloid leukemia—past, present, and future. J Clin Med. 2022;11(3):504. https://pubmed.ncbi.nlm.nih.gov/35159956. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8837075. https://doi.org/10.3390/jcm11030504.
18. Totadri S, Bhatia P, Sreedharanunni S. RUNX1-RUNX1T1-positive acute myeloid leukaemia presenting as bilateral proptosis and multiple cranial nerve palsy. BMJ Case Rep. 2017;2017:bcr2017221402. https://pubmed.ncbi.nlm.nih.gov/28993357. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5652363. https://doi.org/10.1136/bcr-2017-221402.
19. Kondo H, Kanayama T, Matsumura U, et al. Relapsed RUNX1-RUNX1T1-positive acute myeloid leukemia with pseudo-Chediak-Higashi granules. Int J Hematol. 2021;113(5):616-7. https://pubmed.ncbi.nlm.nih.gov/33782817. https//doi.org/10.1007/s12185-021-03141-7.
20. Wei H, Liu X, Wang Y, et al. Optimized clinical application of minimal residual disease in acute myeloid leukemia with RUNX1–RUNX1T1. Exp Hematol. 2021;96:63-72.e3. https://pubmed.ncbi.nlm.nih.gov/33524443 . https://doi.org/10.1016/j.exphem.2021.01.007.
21. Höllein A, Jeromin S, Meggendorfer M, et al. Minimal residual disease (MRD) monitoring and mutational landscape in AML with RUNX1-RUNX1T1: a study on 134 patients. Leukemia. 2018;32(10):2270-4. https://pubmed.ncbi.nlm.nih.gov/29568097. https://doi.org/10.1038/s41375-018-0086-0.
22. Higgins A, Shah MV. Genetic and genomic landscape of secondary and therapy-related acute myeloid leukemia. Genes (Basel). 2020;11(7):749. https://pubmed.ncbi.nlm.nih.gov/32640569. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7397259. https://doi.org/10.3390/genes11070749.
23. Feng Y, Li X, Cassady K, Zou Z, Zhang X. TET2 function in hematopoietic malignancies, immune regulation, and DNA repair. Front Oncol. 2019;9:210. https://pubmed.ncbi.nlm.nih.gov/31001476. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6454012. https://doi.org/10.3389/fonc.2019.00210.
24. Badr P, Elsayed GM, Eldin DN, Riad BY, Hamdy N. Detection of KIT mutations in core binding factor acute myeloid leukemia. Leuk Res Rep. 2018;10:20-5. https://pubmed.ncbi.nlm.nih.gov/30112273. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6092444. https://doi.org/10.1016/j.lrr.2018.06.004.
25. Ghafoor T, Khalil S, Farah T, Ahmed S, Sharif I. Prognostic factors in childhood acute myeloid leukemia; experience from a developing country. Cancer Rep (Hoboken). 2020;3(5):e1259. https://pubmed.ncbi.nlm.nih.gov/33085844. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7941425. https://doi.org/10.1002/cnr2.1259.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 PJP

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.

The Philippine Journal of Pathology is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. Based on works made open access at http://philippinejournalofpathology.org








