Introduction. The National External Quality Assessment Scheme (NEQAS) has been established by the Department of Health–Philippines (DOH) to provide DOH-approved external quality assessment programs, including the Proficiency Test (PT) for Bacteriology to clinical laboratories. The PT for Bacteriology aims to monitor and evaluate laboratory capabilities in the identification of clinically important pathogens through proficiency testing. Since then, participation in the NEQAS has been a requirement for clinical laboratories to obtain a license to operate from the DOH–Health Facilities and Services Regulatory Bureau (HFSRB).
Objective. The objective of this report is to summarize and examine the results of the PT for Bacteriology from 2009 to 2015 and the performances of participating clinical laboratories throughout the Philippines.
Methodology. The Research Institute for Tropical Medicine National Reference Laboratory (RITM-NRL) conducted orientation seminars between 2008 and 2009 to introduce clinical laboratories to the NEQAS. Laboratories submitted their accomplished enrolment forms to RITM–NRL and paid the fees to enroll in the PT. Participating laboratories were required to identify three analytes and perform antimicrobial susceptibility test (AST) on one assigned analyte.
Results. A total of 468 laboratories participated over the seven-year period. The number of participating laboratories obtaining a passing score of 80% and above had significantly increased from 2009 to 2015. Out of the 144 laboratories consistently enrolled over the seven-year period, the proportion of participants with scores of 80% and above had increased. Of the 468 participating laboratories throughout 2009 to 2015, 33.3% were good performers; 6.6% were fair performers; and 60.0% were poor performers.
Conclusion. The increasing number of participating laboratories obtaining passing scores over the years suggests overall improvement of the performance of clinical laboratories in bacteriology. Corrective actions are still needed to address the situation regarding the poor performing laboratories. The assessments done in 2008 and 2013 found that poorly performing laboratories lack trained personnel, resources, and implementation of quality assurance procedures for bacteriological testing.
Key words: laboratory proficiency testing, bacterial identification, antibiotic susceptibility testing
Clinical bacteriology laboratories carry out the detection and isolation of bacterial pathogens for management and surveillance of infectious diseases. They are responsible for detecting antibiotic resistance, identifying outbreak pathogens, and communicating incidences and information concerning infectious diseases to public health authorities. These important tasks are the reason that high quality testing, and accurate and precise results must always be ensured.[1]
The Proficiency Test (PT) for Bacteriology assesses the ability of clinical microbiology laboratories to identify and characterize clinically important bacteria and conduct antimicrobial susceptibility tests. It aims to improve the performances of laboratories and ensure high quality and reliable testing in the field of clinical bacteriology.[2],[3] In other countries, external quality assessment schemes (EQAS) and PTs for clinical microbiology, had been influencing the improvement of the quality of testing and performance of participating laboratories in different time periods.[4],[5],[6],[7] Improvement of laboratory performance can be attained when sources of errors that result to poor performance are identified and addressed.[4],[8],[9]
In order to establish an effective public health laboratory network in the Philippines and streamline its functions, the Department of Health (DOH) of the Republic of the Philippines issued the Department Order No. 393–E s. 2000. It designated the Department of Microbiology of the Research Institute for Tropical Medicine (RITM) as the National Reference Laboratory (NRL) for bacterial enteric diseases, emerging and re-emerging bacterial diseases, and mycology. It also mandates RITM–NRL to maintain a quality assurance program for clinical bacteriology laboratory tests.[10] The Department Administrative Order No. 2007–0027 and Memorandum No. 2009–0086, were then issued by the DOH, which required every clinical laboratory throughout the country to participate in the National External Quality Assessment Schemes (NEQAS) in order to obtain a license to operate (LTO) from the DOH Health Facilities and Services Regulatory Bureau (HFSRB, formerly Bureau of Health Facilities and Services).[11],[12] NEQAS issues DOH-approved external quality assessment programs for bacteriology, parasitology, and mycobacteriology to clinical laboratories by providing a proficiency test that aims to monitor and evaluate the laboratory’s capabilities to identify clinically important pathogens. Thus, the RITM–NRL has been providing annual PTs for bacteriology under the NEQAS for Bacteriology, Parasitology, and Mycobacteriology to clinical laboratories since 2009.
This report summarizes and examines the results of the PT for Bacteriology from 2009 to 2015 and the performances of participating clinical laboratories. The information and analysis were only limited to the scores, regional location, ownership type and accreditation category of 468 participants in the PT throughout the seven-year period and to the data acquired from the assessments of selected participating laboratories.
Baseline assessment of laboratories
A baseline on-site assessment of tertiary clinical laboratories throughout the country was conducted by RITM-NRL in 2008. The assessment aimed to (1) monitor their compliance with the minimum and essential requirements as set by RITM–NRL (Table 1); (2) evaluate their capacity to isolate, identify, and characterize medically important bacterial pathogens; (3) identify their deficiencies, which may result to poor performance in bacteriological testing; (4) educate laboratorians on current microbiological advancements; (5) and promote good laboratory practices. The list of tertiary laboratories provided by HFSRB served as the basis for the number of laboratories to be assessed. The assessment covered laboratory practices on specimen processing: (1) isolation and identification of medically important bacteria; (2) methods for antimicrobial susceptibility testing; (3) internal and external quality assessment practices; (4) use of functional equipment, instruments, culture media, reagents, kits, glassware, and disposables; and (5) waste management.
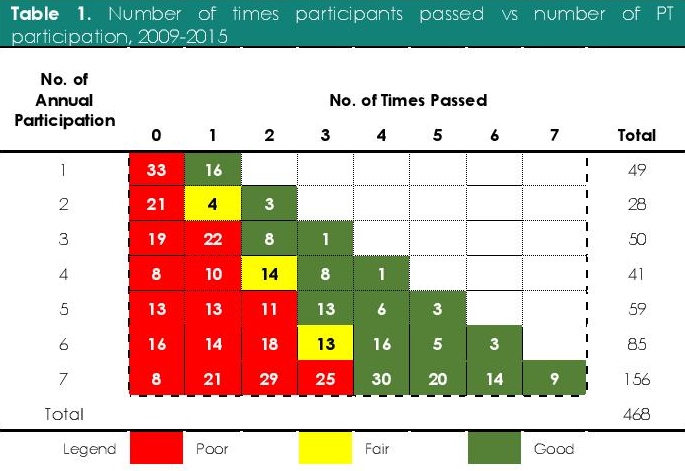
Table 1. Number of times participants passed vs number of PT participation, 2009-2015
Enrolment in the proficiency tests
Since the DOH required clinical laboratories registered under tertiary category in the Center for Health Development (CHD) to participate in the PT event for the first time in 2009, RITM–NRL conducted orientation seminars between 2008 and 2009 to introduce the participants to the NEQAS. Participating laboratories submitted their accomplished enrolment forms through courier, fax, e-mail, or personal delivery and paid the fees before the scheduled testing event to be eligible for the PT. In 2015, DOH required all clinical laboratories under primary, secondary, and tertiary category performing bacteriological testing to participate in the PT.[13]
Materials and analyte preparation
Analyte culture, inoculation, and verification.Clinically-significant bacteria were subcultured from stock cultures in skim milk-tryptone-glucose-glycerol (STGG) media[14] incubated at 36±1°C for 18 to 24 hours. Cultures were examined for purity; assayed through conventional identification methods[15] and commercial identification testing systems: API® (bioMérieux, Marcy-l'Étoile, France) and VITEK® 2 (bioMérieux, Marcy-l'Étoile, France); and compared to ATCC® biological standards (ATCC, Manassas, Virginia, USA) to verify their identities (Table 2). The analytes were inoculated in semi-solid sheep blood agar (SBA)[16] contained in 2 mL-cryogenic vials (Corning Inc., Corning, New York, USA) and incubated at 36±1°C for 18 to 24 hours prior to transport.
Verification of analyte antimicrobial susceptibility. Antimicrobial susceptibility of analytes was verified using BBL™ Sensi-Disc™ Susceptibility Test Discs (Becton, Dickinson, and Co., Sparks, Maryland, USA). The panel of antibiotics used was based on the Performance Standards for Antimicrobial Susceptibility Testing of the Clinical and Laboratory Standards Institute (CLSI, Wayne, Pennsylvania, USA).[17]
Packaging. Analytes sent to participating laboratories were packaged in accordance to the international standard of transporting biohazard materials.18,19 Each vial was sealed with Parafilm M® (Bemis Co. Inc., Oshkosh, Wisconsin, USA), individually wrapped in a paper towel, and placed inside a 100 mm × 150 mm resealable polypropylene resin bags along with other analytes. The wrapped vials were then encased in 600 mL polypropylene canister (Philtop Industries Inc., Valenzuela City, Metro Manila, Philippines) and placed inside a 120 mm × 115 mm × 190 mm corrugated box (Thousand Oaks Packaging Corp., Parañaque City, Metro Manila, Philippines) with the necessary attachments and labels. The package also includes standard proficiency testing guidelines that contain basic information and instructions needed for the handling of the analytes and an answer sheet.
Quality Control of Transport Media and Packaged Analytes. The semi-solid sheep blood agar (SSBA) to be used as transport media was assured for sterility and tested for culture response before use in the PT. Four sets of cultures of each of all the organisms used as analytes in the PT were prepared and each set was subjected to each of the four different treatments. For the first treatment, separate packages containing one set of different analytes were sent to random locations of participating laboratories throughout the country. The selected laboratories were asked to return the sealed package to RITM. Upon return, the analytes were examined for contamination by unwanted organisms and tested for viability through routine culture examination. For the remaining treatments, three sets were incubated at 36±1ºC, 4±2 ºC, and ambient temperature, respectively. Growth was observed after three, five, and seven days. After seven days of incubation, the analytes were subcultured and re-identified through conventional methods and commercial identification systems such as API® and VITEK® 2.
The Bacteriology PT program
Participating laboratories were asked to identify each of the three analytes by means of their routine methods or standard operating procedures. They were also required to perform antimicrobial susceptibility testing on one pre-assigned analyte using the panel of antibiotics recommended by CLSI. After receiving the analytes, participants were given fifteen working days to complete the PT.
The Overall Score, with a perfect rating of 100%, comprised of 75% for organism identification and 25% for AST. RITM–NRL set the passing score to be 80%. A correct identification—with correct binomial name—amounts to 25 points; an acceptable identification—with correct genus but incorrect or unspecified species—amounts to 10 points; an incorrect one amounts to no point. A correct report of antibiotic susceptibility amounts to one point while an incorrect report amounts to no point. The identification of an additional organism would result into an addition to the number of principal organisms in the equation but no addition to the points corresponding to the correct and acceptable identification. This would eventually lead to the reduction of the overall score. The following equation represents the computation for the overall score:

Each of the participating laboratories who had accomplished the PT and submitted their answers to NEQAS received a certificate of participation, a summary of results of its PT performance, and a learning monograph, which recommends standard methods of identifying the analytes and performing AST.
Evaluation of the overall performance of laboratories
Participating laboratories were grouped according to the number of times they enrolled in the annual PTs from 2009 to 2015 and the number of times they got a passing score of 80%. The performances of participating laboratories were classified as “good’, “fair”, and “poor” based on the number of times they passed the annual PT. “Poor performers” refers to participating laboratories who had not met the 80% passing rate for more than 50% of their annual PT participation. “Fair performers” refers to participating laboratories who had met the passing rate in 50% of their annual PT participation. Finally, “good performers” refers to participating laboratories who had passed more than 50% of their annual PT participation.
Assessment of poor performing laboratories
Participating government laboratories that had been consistently obtaining scores below 80% in the 2009–2012 PTs were selected for an on-site reassessment in 2013. Details on the updated training of laboratory personnel; availability of laboratory equipment, reagents, culture media, antibiotics, and glassware; reliance on automated and/or semi-automated systems for identification and AST; use of CLSI Performance Standards for Antimicrobial Susceptibility Testing as guide for AST; availability of ATCC® biological standards; and implementation of quality assurance and control programs were identified.
Statistical analysis
Graphs were generated using Matplotlib version 2.0.0 pyplot module[20] in Python and all statistical analyses were done using SciPy version 0.19.0 scipy.stats module.[21] Friedman ranking test was used to detect differences in the annual scores of consistently enrolled participants in 2009–2015 and Nemenyi test was used as the post hoc test to detect differences between the rankings of annual scores obtained from Friedman test. Multiple comparisons were employed using STAC Web Platform version 1.0.[22]
2008 baseline assessment of laboratories
The HFSRB list included 400 tertiary laboratories throughout the Philippines. Three hundred forty-seven (86.8% of 400) tertiary clinical laboratories capable of doing bacteriological testing were assessed: 25.1% (87/347) of which were located in the National Capital Region; 21.6% (75/347) in Mindanao; 21.0% (73/347) in North Luzon; 16.4% (57/347) in South Luzon; and 15.9% (55/347) in the Visayas. Two hundred seventy-five (79.2% of 347) were private; 70 (20.2% of 347) were government-owned; and two (0.6% of 347) were semi-private. The remaining 53 (13.2% of 400) in the list had already undergone closure or downgrade during the assessment.
In the assessment, only 43.8% (152/347) of the laboratories could perform Gram stain, acid-fast stain, negative stain, and wet mount; 17.3% (60/347) implemented internal quality control procedures for media, reagents, stains, and antibiotic disks; and only 3.2% (11/347) used ATCC biological standards for quality control (Table 1). Only 35.7% (124/347) completed the required essential major equipment and instruments; 5.8% (20/347) as to culture media and supplements for primary isolation; 19.3% (67/347) as to media for biochemical tests; 3.3% (11/347) as to supplements for growth and identification. Notably, 67.7% (235/347) of the laboratories were using human blood in the preparation of blood agar plates, instead of the recommended sheep blood23, which was used by 23.6% (82/347) of the laboratories, or horse blood, which was used by the remaining 6.6% (23/347). Only 17.9% (62/347) had complete sugars for carbohydrate utilization tests and 2.9% (10/347) had complete antibiotics for AST. All in all, only 2.6% (9/347) had complete required essential media and reagents.
Of the 97.4% (338/347) remaining laboratories, 57.9% (201/347) used commercially prepared kits while 24.8% (86/347) used automated systems for identification of bacterial pathogens. For AST, 2.9% (10/347) had complete required essential antibiotics while 15.9% (55/347) of the remaining uses automated systems.
Only 82.9% (288/347) of laboratories were capable of performing AST. Moreover, 76.9% (267/347) of laboratories were performing disk diffusion and 16.7% (58/347) were using automated systems for AST. Only 47.2% (164/347) were using the latest edition of the Performance Standards for Antimicrobial Susceptibility Testing[17] by the Clinical and Laboratory Standards Institute (CLSI), the recommended standard for AST methods and interpretation.
Laboratory participation and performance in 2009–2015
Four hundred sixty-eight participants, comprised of 381 (81.4%) private laboratories and 87 (18.6%) government-owned laboratories, enrolled in the NEQAS for Bacteriology between 2009 and 2015. Of the 450 (96.2% of 468) tertiary category clinical laboratories, 80.7% (363/450) were privately owned, and 19.3% (87/450) were government owned. Eighteen (3.8% of 468) private secondary category laboratories also enrolled in the PTs. The number of participants was highest in the National Capital Region (Figure 1) with 78 (27.3% of 286) participants in 2009, which grew to 112 (29.5% of 403) in 2015.
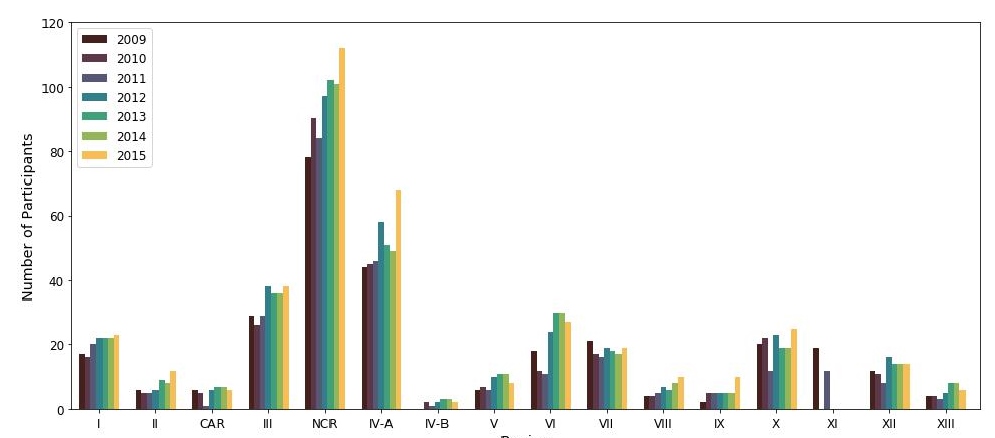
Figure 1. Number of participants from different regions in the Philippines, 2009–2015.
The annual number of participants were the following: 286 (2009), 285 (2010), 264 (2011), 355 (2012), 364 (2013), 360 (2014), and 403 (2015). Scores ranged from 0 to 100 in all years from 2009 to 2015. The mean scores and sample standard deviation per year were: 63.9 ± 28.2 (2009), 70.6 ± 25.0 (2010), 63.1 ± 24.6 (2011), 57.9 ± 28.6 (2012), 60.0 ± 34.3 (2013), 71.1 ± 24.0 (2014), and 75.2 ± 27.3 (2015). The annual median scores were: 71.4 (2009), 75.1 (2010), 68.8 (2011), 60.0 (2012), 72.0 (2013), 72.0 (2014), and 80.0 (2015). The number of participants who obtained scores of 80 and higher increased from 2009 (33.2%, 95/286) to 2015 (57.1%, 230/403). Proportion of passing scores among laboratories participated in PT in bacteriology in 2009–2015 are represented by a line plot (Figure 2).
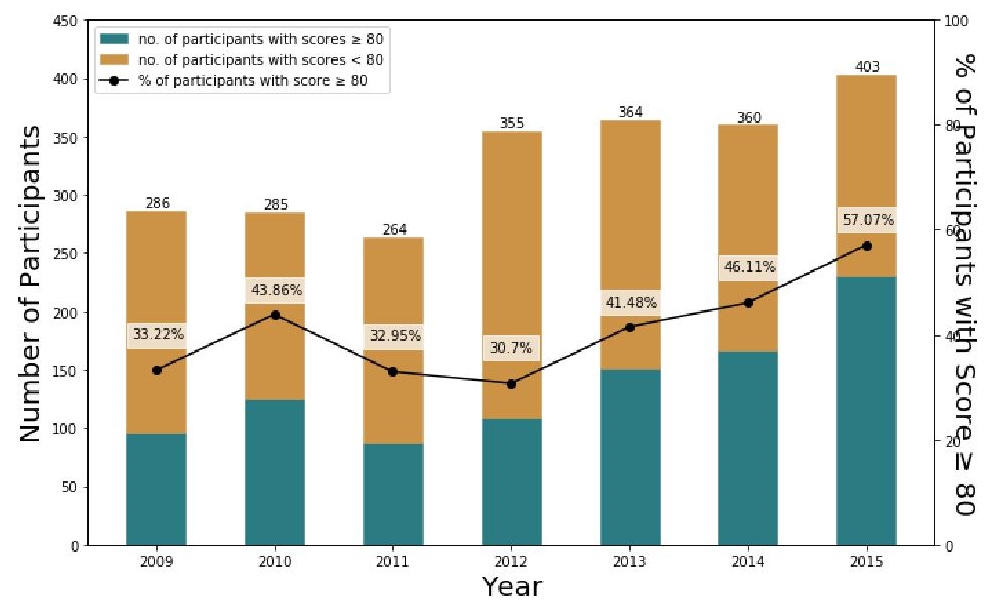
Figure 2. Number of participating laboratories and proportion of passers, 2009–2015; spearman’s ρ=0.821, for both annual number of participants and number of passers; p̂2015 − p̂2009 = 23.9% and χ2 = 38.13.
Out of the 156 (33.3% of 468) good performers, 23.1% (36/156) participants passed all of the annual PTs they enrolled in the span of seven years (Table 1). One hundred eighteen (25.2% of 468) laboratories, on the contrary, never got a passing score in all the PTs, in which they enrolled. Overall, poor performers comprise the most number (60.0%, 281/468) of participating laboratories (Figure 3). A proportion of 40.2% (35/87) government laboratories and 70.4% (247/381) of privately owned laboratories performed poorly in the PT over the seven-year period. While the overall number of participating laboratories and the number of passers were increasing, majority of participating laboratories performed poorly in the PT over the seven years.
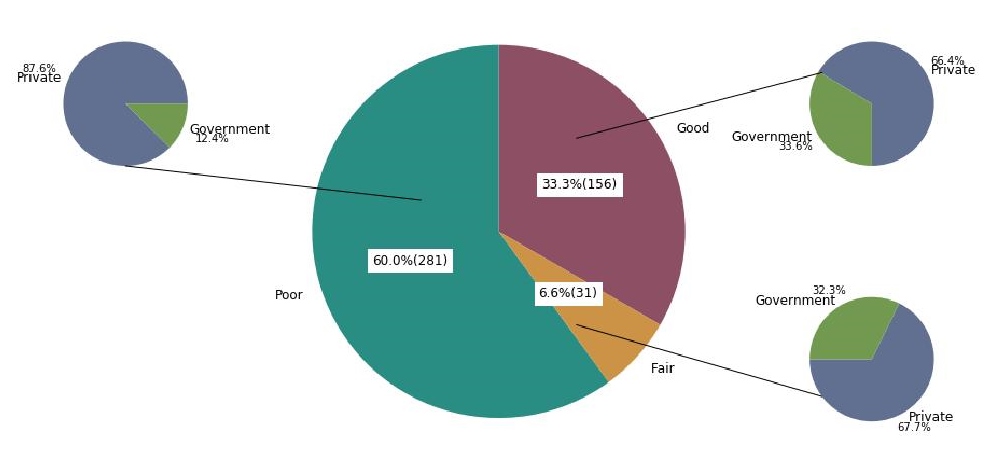
Figure 3. Classification of good, fair, and poor performers, 2009–2015, n=468.
One hundred forty-four laboratories (30.7% of 468) consistently enrolled in the 2009–2015 PTs. The number of laboratories with scores of 80% and above increased from 2009 (37.2%, 50/144) to 2015 (69.4%, 100/144) (Figure 4). Furthermore, the distributions of scores over seven years are negatively skewed, which means the mass of the distribution of scores each year is concentrated in the region of high scores (Figure 5). The majority of distributions of scores in 2010, 2011, 2013, 2014, and 2015 are clustered, resulting to sharper peaks (leptokurtic profiles). The distribution of scores in 2015 holds the highest kurtosis (excess kurtosis = 2.06), which is the measure of the sharpness of the peaks, because of a surge of participants obtaining scores between 90% and 100% (50.7%, 73/144). The highest mean score is found in 2015 (x̅ = 81.7) while the lowest mean score is in 2012 (x̅ = 67.1), followed by the mean score in 2009 (x̅ = 67.6) (Fig. 5). Comparison of annual scores using Friedman test (F = 10.34; P < 0.001) and Nemenyi test as post hoc analysis shows a significant difference between scores in 2009 and in 2015 (Z = 5.55; P < 0.001) with 2015 ranking as the highest (r̅ = 4.95), 2009 as the third from the lowest (r̅ = 3.54), and 2011 as the lowest (r̅ = 3.39) in seven years. The increasing proportion of passers in the 144 consistently enrolled laboratories and the significant difference between scores in 2009 and in 2015 suggest that the performance of the consistently enrolled laboratories had generally improved over the seven-year period.
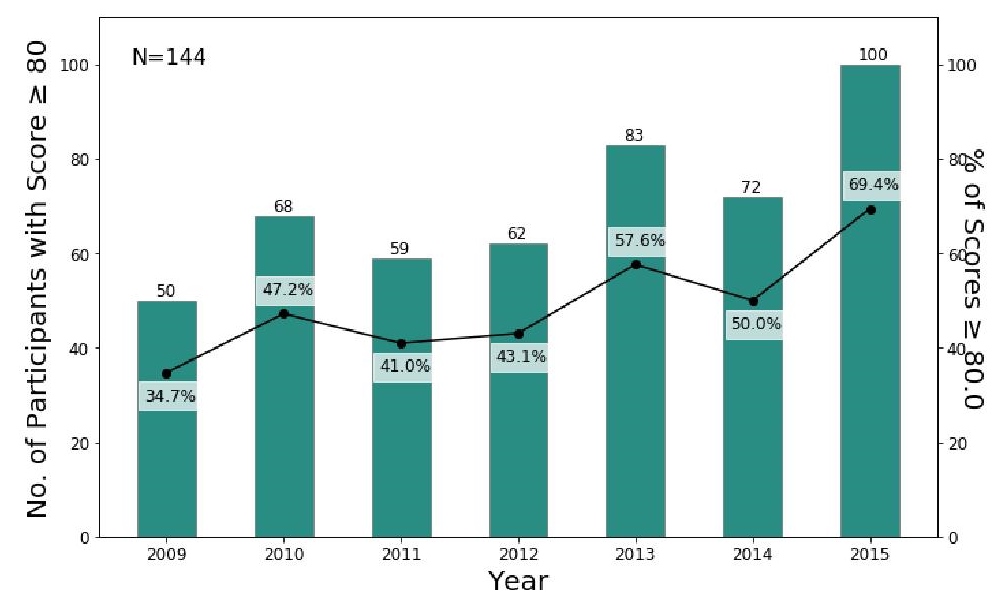
Figure 4. Number and proportion of passers out of laboratories consistently enrolled, 2009–2015; spearman’s ρ=0.821 for the annual proportion of passers of consistently enrolled laboratories.
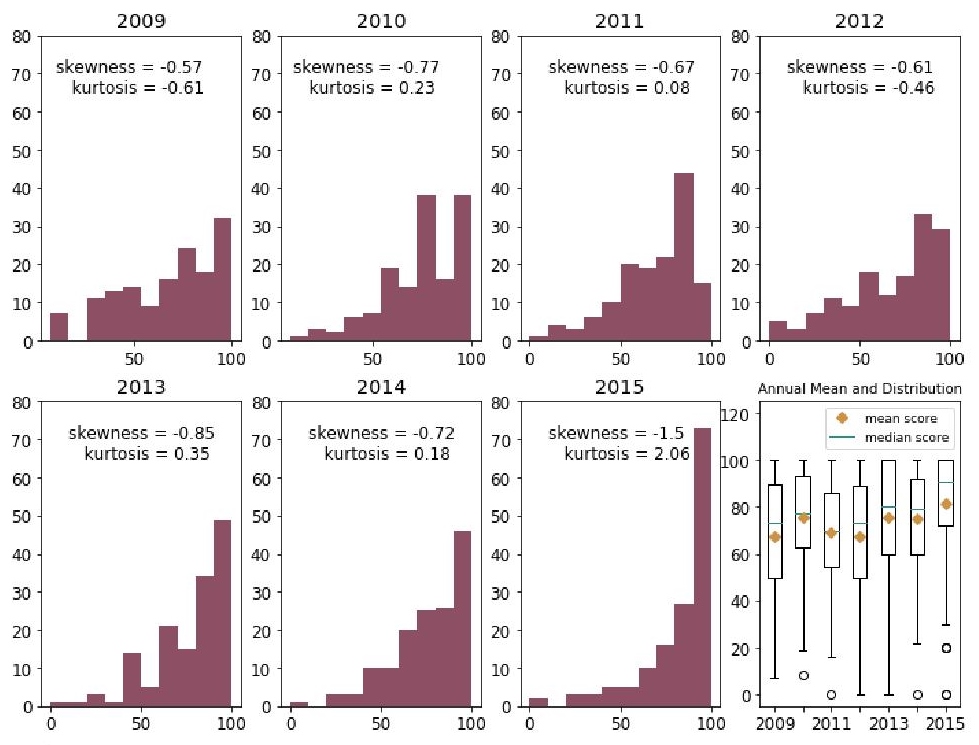
Figure 5. Histogram and box plot of scores plus annual mean scores of 144 laboratories consistently enrolled in the 2009–2015 proficiency tests.
Participation in EQA programs has been proven to improve a laboratory in many ways. It allows the participants to have an idea of their capability and monitor continual improvement, since it generates information that can be used to assess the overall competence and needs of participants.[24]4 It also brings benefits and challenges to the participants, aside from meeting regulatory requirements.[1] The improvement in the performance of bacteriology laboratories in the Philippines in the Proficiency Test provided by NEQAS is comparable to the improvement of laboratories that participated in different EQAS/PT in other countries. In the 1982–1999 Tokyo Metropolitan Government External Quality Assessment Program, an improvement in the performance of independent laboratories in Tokyo, Japan regarding the identification of H. influenzae, MRSA, and some pathogenic enteric bacteria was observed.[6] In the 1992–1996 Swiss External Quality Assessment Scheme in Bacteriology and Mycology, the increasing mean scores of all participants and the number of participating laboratories with high average scores over the four year period reflected the improving performances of participating laboratories.[7] In the United States, the Clinical Laboratory Improvement Amendments of 1988 (CLIA'88) mandate universal requirements for all clinical laboratory-testing sites. This mandate includes the provision of PT that defines laboratory performance. Through PT as one of its tool, CLIA has ensured the adherence of participating laboratories to good clinical practices and improvement in the quality of laboratory tests since 1994.[25] External quality assessment programs are recognized as an effective tool in improving the quality of medical laboratories in Europe.[26] Further improvements are being considered to their existing EQA programs such as accreditation of schemes and further integration to information technology[27] that can also be applied in the Philippines.
Quality control of packaged analytes and the analyte material
PTs and EQAS in other countries use either simulated clinical samples or lyophilized cells as analytes. Simulated clinical samples (e.g. nose swabs, artificial feces, simulated spinal fluid, etc.) provide clinically relevant and realistic challenges yet they require appropriate facilities and technology, and arduous effort.[28] Lyophilization of cells for transport is straightforward and it protects cells from degradation; however, the subsequent processes of reconstitution and multiple passages do not demonstrate clinical relevance and realism. It completely lacks resemblance to clinical specimens.[4] Furthermore, artificial handling and significant matrix effects can affect the growth, colony morphology, and nutrient metabolism of organisms, thus affecting proper identification and characterization of analytes.[28] In this PT, the use of semi-solid SBA as matrix was proven to be effective in ensuring the viability of organisms during transport, through quality control. All of the isolates sent for quality control throughout 2009 to 2015 were observed to be contaminant-free and viable.
Identification and AST
Bacteriology laboratories in the Philippines used conventional methods, manual commercial kits, and automated systems for the characterization and identification of clinically significant pathogens. The organisms identified with least difficulty were E. coli, S. aureus, MRSA, C. albicans, S. enterica sv. Typhi, and P. aeruginosa. In contrast, the six organisms identified with highest difficulty were A. lwoffii, C. tropicalis, M. catarrhalis, E. aerogenes, H. influenzae, and S. pneumoniae (Table 2). Additionally, the antibiotic susceptibilities of S. pneumoniae, E. cloacae, and H. influenzae were the most difficult to determine (Table 3).
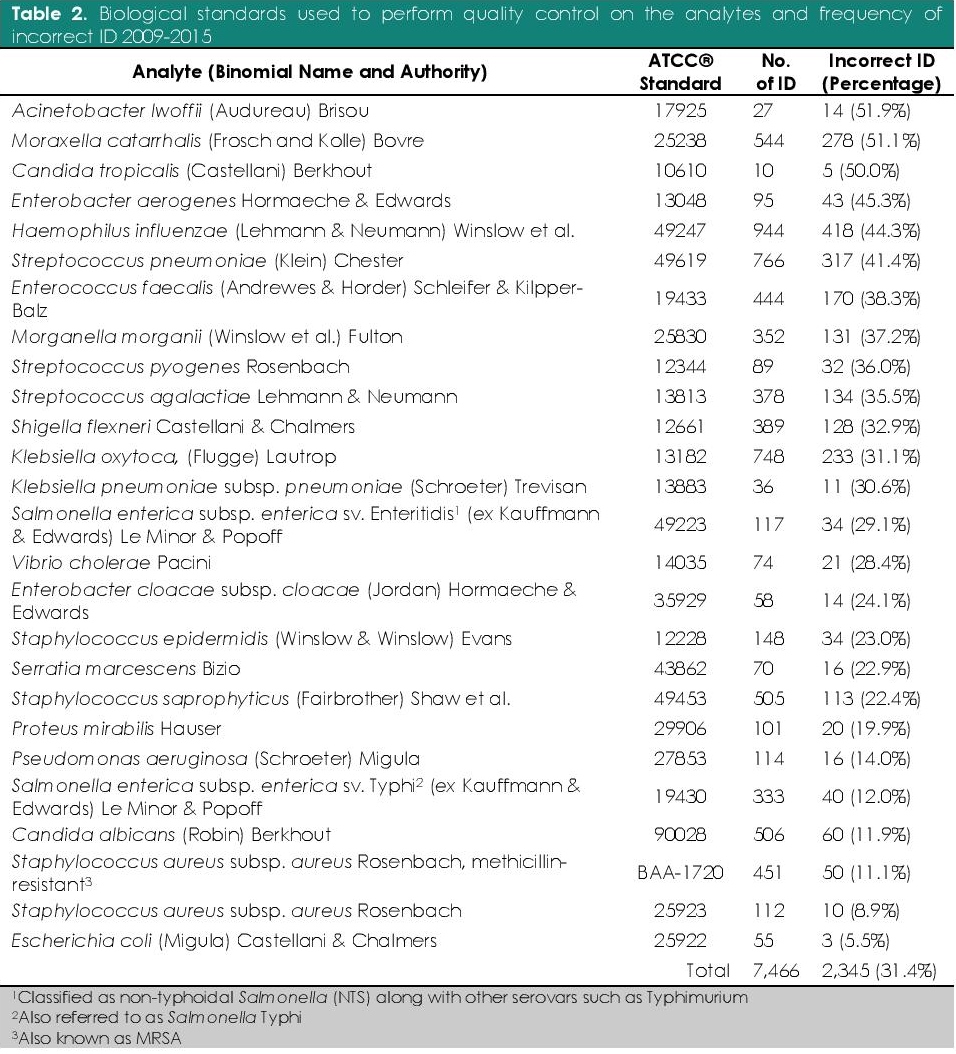
Table 2. Biological standards used to perform quality control on the analytes and frequency of incorrect ID 2009-2015
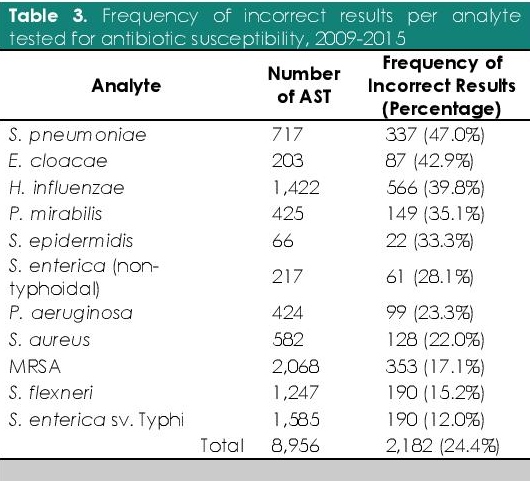
Table 3. Frequency of incorrect results per analyte tested for antibiotic susceptibility, 2009-2015
The organisms identified with least difficulty are usually distinguished with colony examination and few straightforward biochemical tests. S. aureus is identified as Gram-positive coccus, which is positive for catalase and coagulase tests. Salmonella enterica sv. Typhi can be distinguished from nontyphoidal Salmonella with its distinct biochemical characteristics: it is citrate-negative, ornithine-negative, and mucate-negative; it yields alkaline products on aerobic environment, acidic products on anaerobic environment; and it weakly produces hydrogen sulfide gas in triple sugar iron (TSI) agar. Moreover, S. enterica sv. Typhi can be differentiated from other Salmonella serotypes through serological tests based on the antigenic properties of the somatic (O:9), flagellar (H-d), and capsular (Vi) antigens. The identity of the yeast, C. albicans, can be confirmed when it produces germ tube during germination in horse serum at 37°C, and terminal chlamydospores on hyphae or pseudohyphae during growth in corn meal agar at 25°C. E. coli and P. aeruginosa can be identified through examination of colony morphology in MacConkey agar and a few basic biochemical tests.[15]
Some of the organisms identified with highest difficulty, A. lwoffii and M. catarrhalis, are nonfermentative Gram-negative bacteria that belong to the Moraxellaceae family. Acinetobacter species are oxidase-negative, catalase-positive, indole-negative coccobacillary bacteria. A. lwoffii can be differentiated from other Acinetobacter species through carbon assimilation tests.[15] Moraxella species, on the other hand, are oxidase-positive, catalase-positive, indole-negative coccoid or coccobacillary bacteria. They can be differentiated from the similarly oxidase-positive and catalase-positive Neisseria species through examination of the colonies in agar: Moraxella colonies may be pushed intact across the plate with a loop like a hockey puck; or through DNase and tributyrin tests. M. catarrhalis can be distinguished from other Moraxella species through its ability to reduce nitrate and nitrite and its inability to alkalinize acetate and acidify ethylene glycol. Enterobacter species belong to the Enterobacteriaceae family. E. aerogenes can be differentiated from other Enterobacter species through lysine decarboxylase, arginine dihydrolase, ornithine decarboxylase, and carbohydrate fermentation tests.[15] C. tropicalis, a non-albicans Candida (NAC) yeast species,[29] is germ tube-negative. It can be distinguished from other Candida species through microscopic examination of morphological features of the yeast on cornmeal agar; through carbohydrate assimilation tests; and through carbohydrate fermentation tests.[15]
In the 2008 assessment, majority (66.7%, 231/347) of the laboratories assessed used human blood agar in the isolation, detection, and characterization of clinically important bacterial pathogens. These pathogens, especially fastidious organisms such as S. pneumoniae and H. influenzae, are less likely to be detected when using human blood agar for culture since antibodies and residual antibiotics in human blood may inhibit the growth of bacterial isolates. It will also result to pathogens producing incorrect or varying hemolysis on the blood plate agar which is one of the characteristics critical when identifying a microorganism. Instead, trypticase soy agar plate with 5% defibrinated sheep, goat, rabbit, or horse blood is recommended for the preparation of blood agar plate and as primary culture plate media for bacterial pathogens.[23] Streptococcus pneumoniae and H. influenzae are fastidious organisms, which grow best at 36±1°C with around 5%–10% carbon dioxide or in a candle jar.S. pneumoniae can be differentiated from other streptococci through characterization of colonies on blood agar plates; optochin test; and bile solubility test using sodium deoxycholate. H. influenzae, on the other hand, requires hemin (X factor) and nicotinamide adenine dinucleotide (V factor) for growth; thus, the chocolate agar plate, which contains both factors, is used as the standard growth medium.[15],[23] In addition, S. pneumoniae and H. influenzae must be grown in Mueller-Hinton agar (MHA) supplemented with additional growth factors for AST. MHA with 5% sheep blood is the medium required for AST of S. pneumoniae by disk diffusion. Plates must be incubated at 35±2°C with 5% CO2 for 20–24 hours. Haemophilus test medium (HTM; comprised of MHA plus hematin, nicotinamide adenine dinucleotide, and yeast extract) is required for the AST of H. influenzae by disk diffusion, and plates must be incubated at 36±1°C with 5%–10% CO2 for 16–18 hours.[17]
Organisms that are difficult to identify require additional tests, equipment, media, supplements, and reagents. Failure to identify organisms that require more complex bacteriological testing can be attributed to the large proportion of laboratories lacking minimum and required essential media and reagents, such as sugars for carbohydrate utilization tests and amino acids for amino acid metabolism tests, based on the result of the 2008 assessment. Also, failure to determine the correct antibiotic susceptibility of bacteria through disk diffusion can be attributed to the large proportion of laboratories that were not using the latest edition of the Performance Standards for Antimicrobial Susceptibility Testing by the CLSI in 2008. Laboratories are expected to correctly interpret the zone diameter breakpoints in disk diffusion according to the updated CLSI standards. Moreover, accurate AST results cannot be achieved with the lack of antibiotics, media (e.g. MHA, etc.), supplements (e.g. non-human mammalian blood, hematin, etc.), and equipment (e.g. CO2 incubators or candle jars, etc.).
2013 Assessment of poor performing government laboratories for bacteriology
Only 31 government-owned laboratories that performed poorly in the 2009–2012 PTs were assessed. Two (6.5% of 31) were found to be non-functional due to a lack of budget. Only 12.9% (4/31) of the laboratories had personnel with updated training; 45.2% (14/31) used ATCC biological standards for quality control; and only 29.0% (9/31) implemented SOPs and quality control of culture media, antibiotics, and equipment. Only 9.7% (3/31) had complete essential equipment; 9.7% (3/31) had complete essential culture media and antibiotics for AST; and none (0/31) of the laboratories had complete essential reagents. On the other hand, 83.9% (26/31) had complete essential glassware. Overall, majority of the assessed laboratories did not meet the minimum and essential requirements, except for having complete essential glassware.
Thirteen (41.9% of 31) laboratories use automated and semi-automated equipment. Six laboratories (19.4% of 31) were using VITEK® 2; four (12.9% of 31) were using API® identification testing kits; and three (9.7% of 31) were using BBL™ Crystal™ Identification Systems (Becton, Dickinson and Co. Diagnostic Systems, Sparks, Maryland, USA). Automated and semi-automated systems require freshly grown isolates within 24 hours as its test template; hence, laboratorians still need the basic materials, equipment, and skills to culture and isolate medically important bacteria and fungi. Laboratorians also need to check the viability and density measurement of the organisms to be tested for AST since automated systems which conducts its AST based on broth microdilution testing method, generally require more than 10[5] viable cells.[30] Moreover, failure to assign an identification to an organism as a result of low discrimination and discordant identifications by automated and semi-automated systems, still warrants supplemental and confirmatory testing by conventional methods,[31] which require the necessary materials included in the minimum and essential requirements for bacteriological testing.
The baseline assessment of the 347 tertiary laboratories conducted in 2008 and the assessment of 31 poorly performing government laboratories conducted in 2013 found almost similar findings: poor performance was due to poor compliance to the recommended minimum and essential requirements set by RITM–NRL. Both assessments had recommended poor performing laboratories re-training of laboratory personnel; acquisition of the unavailable media, supplements, reagents, and instruments; management on quality control procedures for media, reagents, antibiotics, and stains; use of the current CLSI standards for AST; and review of skills for bacteria culture, isolation and detection.
More actions are still needed to have a better idea of the current state of clinical bacteriological testing in the country and how it affects the results that are being produced. A wider reassessment, that will aim to include all of the participating laboratories, needs to be carried out urgently and regularly in order to identify factors that lead to poor performance and, likewise, ensure high quality testing and accurate reporting of results. Other information can also be acquired during the reassessment such as compliance or deficiencies in skills, training, resources, and implementation of quality assurance procedures. Further data on the methods used (conventional, commercial or semi-/fully automated system) and how it affects the performance of the laboratory can be investigated. The NEQAS program was established to improve the capacity of participating laboratories in producing quality results. This will lead to the elevation of the state of clinical bacteriology in the country and produce quality service for the Filipino people. In aid of this vision, the RITM together with the DOH offers trainings and assistance that can help the participating laboratories in reaching this goal. Other ideas can be explored to attain this goal such as creating a network among the laboratories that may enable them to share knowledge and resources to improve each other’s performance and capabilities. The data that will be gathered in the following years and succeeding plans will be included in future reports and studies.
An increasing number of participating laboratories had participated in the PT for Bacteriology and the performances of those consistently enrolled had generally improved over 2009–2015. Moreover, a comparison between distributions of scores over the seven-year period has shown an increase in the number of participating laboratories obtaining high to perfect scores. This progress demonstrates that the NEQAS for Bacteriology had improved the quality and reliability of their methods in identifying bacterial pathogens and detecting antibiotic resistance.
In contrast, the large portion of poorly performing laboratories needs to be addressed. The baseline assessment in 2008 and assessment of poor performers in 2013 identified the deficiencies of clinical microbiology laboratories in skills, training, resources, and implementation of quality assurance procedures. A nationwide reassessment of participating laboratories needs to be carried out urgently and regularly in order to identify factors that lead to poor performance and, likewise, ensure high quality testing and accurate reporting of results.
The authors thank the medical laboratory scientists of the National Reference Laboratories (NRL) under the RITM Department of Microbiology: Josefina Geronimo and Rosa Mate of NRL–Bacterial Enteric Diseases (BED); Rizalina Navarro and Pearl Joy Nazareno of NRL–Mycology; Salvacion Rosario Galit of NRL–Emerging and Re-emerging Bacterial Diseases (ERB); and Gloria Reclusado of NRL–Invasive Bacterial Vaccine Preventable Diseases (IBVPD). The authors likewise thank the following administrative staff: Armando Martinez and Sheila Joy Gonzaga of the RITM–NEQAS for Bacteriology, Parasitology, and Mycobacteriology; and Natalie Del Mundo of the RITM Department of Microbiology.
All authors certified fulfillment of ICMJE authorship criteria.
The authors declared no conflict of interest.
The 2009–2012 Proficiency Tests for Bacteriology were funded by the Health Facilities and Services Regulatory Bureau and the Health Facilities Development Bureau—both under the Department of Health of the Republic of the Philippines.
[1] Stang HL, Anderson NL. Use of proficiency testing as a tool to improve quality in microbiology laboratories. Clin Microbiol Newsl. 2013;35(18):145-52. PubMed Central NIHMSID: NIHMS742893 DOI.
[2] WHO Southeast Asian Regional Office. Quality assurance in bacteriology and immunology. New Delhi: World Health Organization, 2012. http://apps.searo.who.int/PDS_DOCS/B4871.pdf?ua=1.
[3] Jones RN, Glick T, Sader HS, et al. Educational antimicrobial susceptibility testing as a critical component of microbiology laboratory proficiency programs: American Proficiency Institute results for 2007–2011. Diagn Microbiol Infect Dis. 2013;75(4):357-60. PubMed DOI.
[4] Snell JJ, De Mello JV, Gardner PS. The United Kingdom national microbiological quality assessment scheme. J Clin Pathol. 1982;35(1):82-93. PubMed PubMed Central.
[5] Whitby JL, Black WA, Richardson H, Wood DE. System for laboratory proficiency testing in bacteriology: organisation and impact on microbiology laboratories in health care facilities funded by the Ontario Government. J Clin Pathol. 1982;35(1):94-100. PubMed PubMed Central.
[6]Kumasaka K, Kawano K, Yamaguchi K, et al. A study of quality assessment in clinical microbiology performance of independent laboratories in Tokyo: 18-year participation in the Tokyo Metropolitan Government External Quality Assessment Program. J Infect Chemother. 2001(2);7:102-9. PubMed DOI.
[7] Siegrist HH, Pünter-Streit V, von Graevenitz A. The Swiss External Quality Assessment Scheme in Bacteriology and Mycology 1992-1996. Accreditation Qual Assur. 1998;3(5):203-07.
[8] Frean J, Perovic O, Fensham V, et al. External quality assessment of national public health laboratories in Africa, 2002–2009. Bull World Health Organ. 2012(3);90:191-9A. PubMed Central DOI.
[9] Snell JJS. Problems in susceptibility testings—findings of UK NEQAS for microbiology. J Antimicrob Chemother. 1994;33:1-4. https://academic.oup.com/jac/article/33/1/1/717194.
[10] Romualdez A. Department Order No. 393-E s. 2000: Designation of National Reference Laboratories and transfer of corresponding equipment, instruments, supplies, specimens, records from the Bureau of Research and Laboratories to the designated National Reference Laboratories. November 2000. http://lcp.gov.ph/images/Dept_Order_393E_s2000.pdf.
[11] Duque F. Administrative Order No. 2007-0027: Revised rules and regulations governing the licensure and regulation of clinical laboratories in the Philippines. August 2007.
[12] Duque F. Department Memorandum No. 2009-0086: Implementation of External Quality Assessment Program as a regulatory requirement for licensing of clinical laboratories. February 2009.
[13] Lutero N. Department Memorandum No. 2009-0086-B: Amendment to Department Memorandum No. 2009-0086-A entitled, “Implementation of External Quality Assessment Program as regulatory requirement for licensing of clinical laboratories.” September 2014. http://lcp.gov.ph/images/Dept_Memo_2009_0086B.pdf.
[14] O’Brien KL, Bronsdon MA, Dagan R, et al. Evaluation of a medium (STGG) for transport and optimal recovery of streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol. 2001;39(3):1021-4. PubMed PubMed Central DOI.
[15] Pfaller MA, Richter SS, Funke G, et al., eds. Manual of Clinical Microbiology, 11th Edition. American Society of Microbiology, 2015.
[16] Atlas RM, Snyder JW. Handbook of media for clinical and public health microbiology. Boca Raton: CRC Press, Taylor and Francis Group, 2014.
[17] Patel JB, Weinstein MP, Eliopoulos GM, et al. Performance standards for antimicrobial susceptibility testing. 27th ed. Clinical and Laboratory Standards Institute, 2017.
[18] Dangerous goods regulations (iata--resolution 618 attachment “a”): effective 1 January-31 December 2015. Montreal: Intl Air Transport Assn, 2014.
[19] Chosewood LC, Wilson DE, eds. Biosafety in microbiological and biomedical laboratories. 5th ed. Washington: U.S. Department of Health and Human Services Public Health Service Centers for Disease Control and Prevention National Institutes of Health, 2010.
[20] Droettboom M, Caswell TA, Hunter J, et al. Matplotlib/Matplotlib: V2.0.0. January 2017.
[21] Jones E, Oliphant T, Peterson P, et al. SciPy: Open Source Scientific Tools for Python. 2001.
[22] Rodriguez-Fdez I, Canosa A, Mucientes M, Bugarin A. STAC: A web platform for the comparison of algorithms using statistical tests. In: IEEE; 2015:1-8.
[23] Castillo D, Harcourt B, Hatcher C, et al. Laboratory Methods for the Diagnosis of Meningitis Caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenza, 2nd ed. Geneva, Switzerland: World Health Organisation Press, 2011.
[24] Barbé B, Yansouni CP, Affolabi D, Jacobs J. Implementation of quality management for clinical bacteriology in low-resource settings. Clin Microbiol Infect. 2017;23:426-33. PubMed DOI.
[25] Ehrmeyer SS, Laessig RH. Has compliance with CLIA requirements really improved quality in US clinical laboratories? Clin Chim Acta. 2004;346(1):37-43. PubMed DOI.
[26] Crucitti T. National External Quality Assessment Schemes for microbiology, parasitology, and virology in Europe. Accreditation Qual Assur. 2001;6(8):379-81.
[27] Libeer JC. Role of external quality assurance schemes in assessing and improving quality in medical laboratories. Clin Chim Acta. 2001;309(2):173-7. PubMed.
[28] Noble MA. Advances in microbiology EQA. Accreditation Qual Assur. 2002;7(8-9):341-4.
[29] Krcmery V, Barnes AJ. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J Hosp Infect. 2002;50(4):243-60. PubMed DOI.
[30] van Belkum A, Dunne WM. Next-Generation Antimicrobial Susceptibility Testing. J Clin Microbiol. 2013;51(7):2018-24. PubMed PubMed Central DOI.
[31] Kiska DL, Kerr A, Jones MC, et al. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34(4):886-91. PubMed PubMed Central.