Key words: Alocilja magnetic nanoparticles, E. coli O157:H7, TEM images, cell capture
Escherichia coli O157:H7 (EcO157) is a notorious foodborne pathogen known to cause bloody diarrhea and can even lead to death. Current detection methods, though highly sensitive, are lengthy and labor-intensive thus an alternative that is simple, rapid, low-cost and equally sensitive is necessary. Hence, an enabling method is the use of functionalized Alocilja magnetic nanoparticles (AMN), known to have high surface reactivity and can easily capture target biomolecules without the use of antibodies, such as microbial cells, in crude samples by means of a magnet. AMN, patented after its inventor Dr. Evangelyn Alocilja, is composed of iron oxide/glycan core/shell structure with an average size of 180-450 nm and with superparamagnetic properties. AMN has been reported to capture Salmonella enterica, Bacillus cereus and Mycobacterium smegmatis without the use of antibodies or peptides.[1],[2],[3],[4]
In this preliminary work, our group tested the ability of AMN to capture a model organism, E. coli O157:H7 isolates provided by the National Institute of Molecular Biology and Biotechnology (BIOTECH), University of the Philippines Los Baños (UPLB) Accession No. 10308 from dairy cattle (Bos taurus L.) feces.[5] O157 latex agglutination test (Oxoid Ltd., Thermo-Fischer Scientific, UK) was used to confirm the identity of the isolates. This strain was characterized through polymerase chain reaction (PCR) to carry shiga toxin-producing genes such as stx1 which causes severe and fatal disease.
To demonstrate this, 100 µL of AMN solution (5 mg/mL) was added to a tube containing 1 mL of a 5-hr old pure EcO157 broth culture (with an initial population of ~108 CFU/mL). The mixed solution was then serially diluted and spot plated on Tryptic Soy Agar (TSA) (18-24 hours, 37°C) to determine the cell population prior to AMN capture. A serially diluted tube was sealed and gently mixed by inversion followed by a 5-min incubation at ambient conditions to allow conjugation of AMN to the cells. Magnetic separation for 1 min was performed to immobilize the resultant AMN-EcO157 complex. The supernatant was aspirated and discarded, and the AMN-EcO157 complex was resuspended in phosphate buffered solution (pH = 7.4). Spot plating on TSA was done to determine the population of bound cells. The percent cell capture efficiency (%CCE) was calculated by dividing the log10 of CFU/mL of captured cells over the log10 of CFU/mL before capture. The average %CCE of AMN towards pure EcO157 is 88.1±1.5 at pH = 7.4.
In order to visualize the capture of EcO157 by AMNs, the residual samples were further analyzed through transmission electron microscopy (TEM). Samples (50 to 100 µL) were dropped onto a 200 mesh formvar coated EM grids for 3 minutes at 25°C and drained. Grids were placed in a pre-labeled polystyrene petri dish lined with Whatmann filter paper and dried in electric desiccator cabinet overnight and examined using the JEOL JEM-1220 TEM under direct magnification of 2000x-3000x at 100 kV.
TEM micrographs confirmed conjugation between the AMPs and EcO157. Figure 1 shows that AMPs were effective in capturing the model pathogen by surrounding the entire cell. Aggregates of EcO157 were seen to interface with the AMNs in Figure 2. Also, it can be noted here that AMNs tend to heap up when far from biotic cells. In Figure 3, bacterial capture is evident despite the presence of a few AMNs.
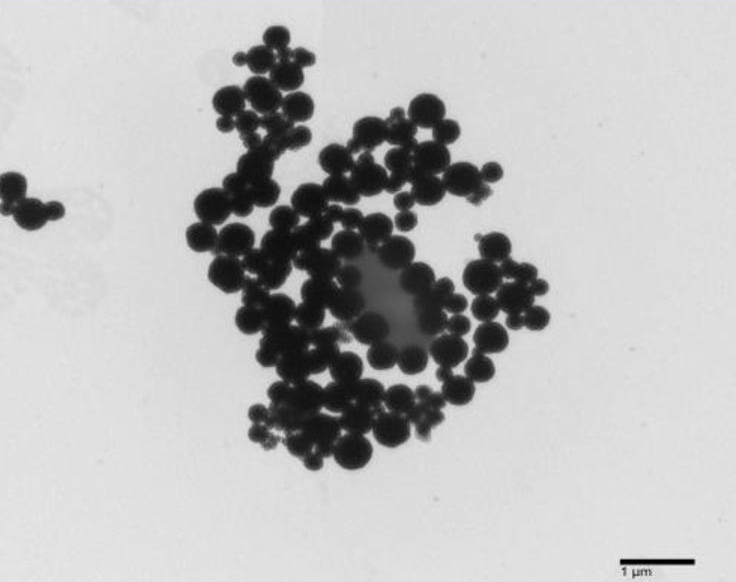
Figure 1. An AMN-EcO157 complex (TEM, mag: 2000x).
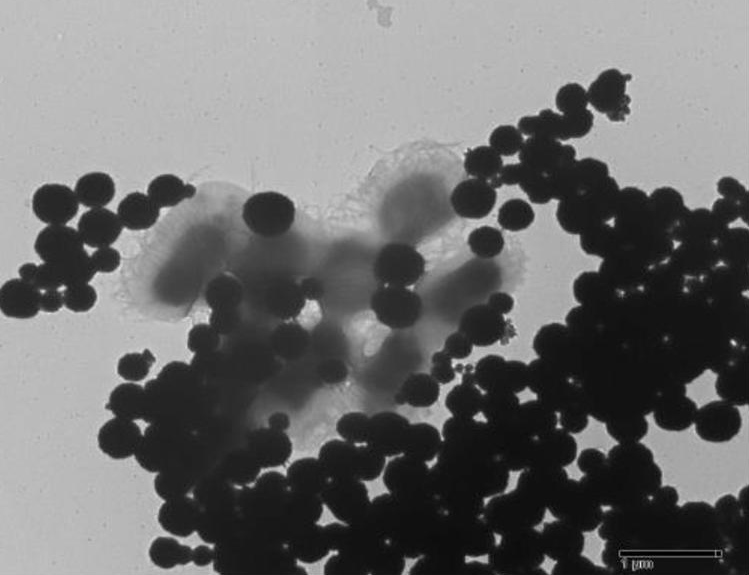
Figure 2. AMNs capturing aggregates of Ec0157 (TEM, 2500x).

Figure 3. EcO157 captured by few AMNs (TEM, 3000x).
Such capture of AMNs to EcO157 can be exploited to develop a potential for simple, centrifuge-free, preconcentration step for further downstream processing such as gDNA extraction, and sensitive DNA-based biosensor detection. Further studies are recommended to optimize test and apply AMN to capture cells from crude samples. The capture of AMNs to pathogens are governed by a number of biological phenomena, such as microbial adhesion, cell surface hydrophobicity, aggregation , biofilm formation, and surface to surface mediation such as hydrodynamic, Lifshitz-Van der Walls, electrostatic, acid-base and hydrophobic interaction forces.[6],[7][8] AMN in solution increases particle density and surface area which promotes higher Brownian movement of the bacteria and AMNs.[6],[7][8] Moreover, ionic and electrostatic interaction between the positively charged AMNs and negatively charged bacterial cell surface adds to the cohesive dynamics of the interaction.[9],[10] Carbohydrate-binding proteins on bacterial cell wall also promotes aggregation and conjugation to nanoparticles.[11]Compounding all these interactive forces contributes to the synergy of cell capture by AMNs.
We thank Dr. Socorro P. Lupisan, Dr. Amado O. Tandoc III, Dr. Ma. Cecilia G. Ama, Dr. Alpha Grace Cabic, Ms. Rocelle Marie Agero (Research Institute for Tropical Medicine); Dr. Rosario G. Monsalud, Dr. Francisco Elegado, David Joram Mendoza, Ma. Theresa Jonna A. Atienza, Ma. Theresa Perez (BIOTECH, UP Los Baños).
All authors certified fulfillment of ICMJE authorship criteria.
The authors declared no conflicts of interest.
This study was funded by the Research Institute for Tropical Medicine and is a recipient of the UP System Emerging Interdisciplinary Research (EIDR) Grant.
[1] Alocilja EC. Laboratory Research Division Brown Bag Presentation: nanotechnology for rapid diagnosis of infectious diseases. Metro Manila, Philippines: Research Institute for Tropical Medicine; 2017.
[2] Zhang D, Carr DJ, Alocilja EC. Fluorescent bio-barcode DNA assay for the detection of Salmonella enterica serovar Enteritidis. Biosens Bioelectron 2009;24(5):1377–81. PubMed DOI.
[3] Vetrone SA, Huarng MC, Alocilja EC. Detection of Non-PCR Amplified S. enteritidis genomic DNA from food matrices using a gold-nanoparticle DNA biosensor: a proof-of-concept study. sensors 2012;12(12):10487–99. PubMed Central
[4] Pal S, Ying W, Alocilja EC, Downes FP. Sensitivity and specificity performance of a direct-charge transfer biosensor for detecting Bacillus cereus in selected food matrices. Biosyst Eng. 2008;99(4):461–8. DOI.
[5] Rundina MCN. Isolation and characterization of Shiga (Vero) Toxin-Producing Escherichia coli O157:H7 in dairy cattle (Bos Taurus) from selected farms in Luzon, Philippines. College, Laguna, Philippines: University of the Philippines, Los Baños; 2004 [master’s thesis].
[6] Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol 2014;4:112. PubMed Central DOI.
[7] Xuan Y, Li Q, Hu W. Aggregation structure and thermal conductivity of nanofluids. AIChE J 2003;49(4):1038–43. DOI.
[8] Boks NP, Norde W, van der Mei HC, Busscher HJ. Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology. 2008;154(10):3122–33. PubMed DOI.
[9] van Loosdrecht MC, Lyklema J, Norde W, Zehnder AJB. Bacterial adhesion: a physicochemical approach. Microb Ecol. 1989;17(1):1–15. PubMed DOI.
[10] van Loosdrecht MC, Lyklema J, Norde W, Schraa G, Zehnder AJ. Electrophoretic mobility and hydrophobicity as a measured to predict the initial steps of bacterial adhesion. Appl Environ Microbiol. 1987;53(8):1898–901. PubMed Central.
[11] dela Fuente JM, Penadés S. Glyconanoparticles: types, synthesis and applications in glycoscience, biomedicine and material science. Biochim Biophys Acta. 2006;1760(4):636–51. PubMed DOI.